This area is for writing your GenoCon2 Report. The guidelines below are for writing a 6 section report. An image should also be included.
Title: Design for combining HPS (3-hexulose-6-phosphate synthase) and PHI (6-phospho-3-hexuloisomerase)
Name: RIKEN GenoCon
Affiliation: RIKEN The Institute of Physical and Chemical Research
Date: 2012-10-10
Abstract
Formaldehyde is known as a harmful substance to the human body and cause sick house syndrome. If there was a plant which is able to absorb formaldehyde and remove it effectively, we might be able to prevent sick house syndrome by putting the plant in the room. However, general plants have little function to absorb formaldehyde. In this design, we have used JavaScript to design DNA sequence to add such a function to a model plant.The resulting design is efficient, simple, and easy to understand example of JavaScript coding to construct a DNA sequence created by reverse transcription. There are many possibilities to improve on this design.
Introduction
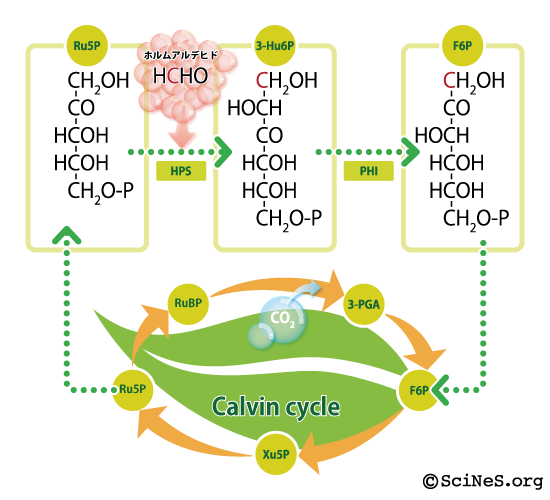
Research groups led by Professor Izui of Kinki University, Professor Sakai, and Professor Kato (currently Professor of Kyoto Gakuen University) of Kyoto University are actually trying to create such dreamlike plants. They found out that by inserting two HCHO decomposing bacterium genes into a plant (Arabidopsis thaliana), the plant got a function to remove HCHO in the air. The research teams re-design a plant pathway by adding two genes. (See details of the research)
Method
Overview
-
Obtain the HPS protein sequence
-
Reverse-translate the obtained HPS protein sequence to DNA sequence
-
Obtain PHI protein sequence
-
Reverse-translate the obtained PHI protein sequence to DNA sequence
-
Combine those two DNA sequences
-
Register the combined DNA sequences in RIKEN SciNetS

Design Flow
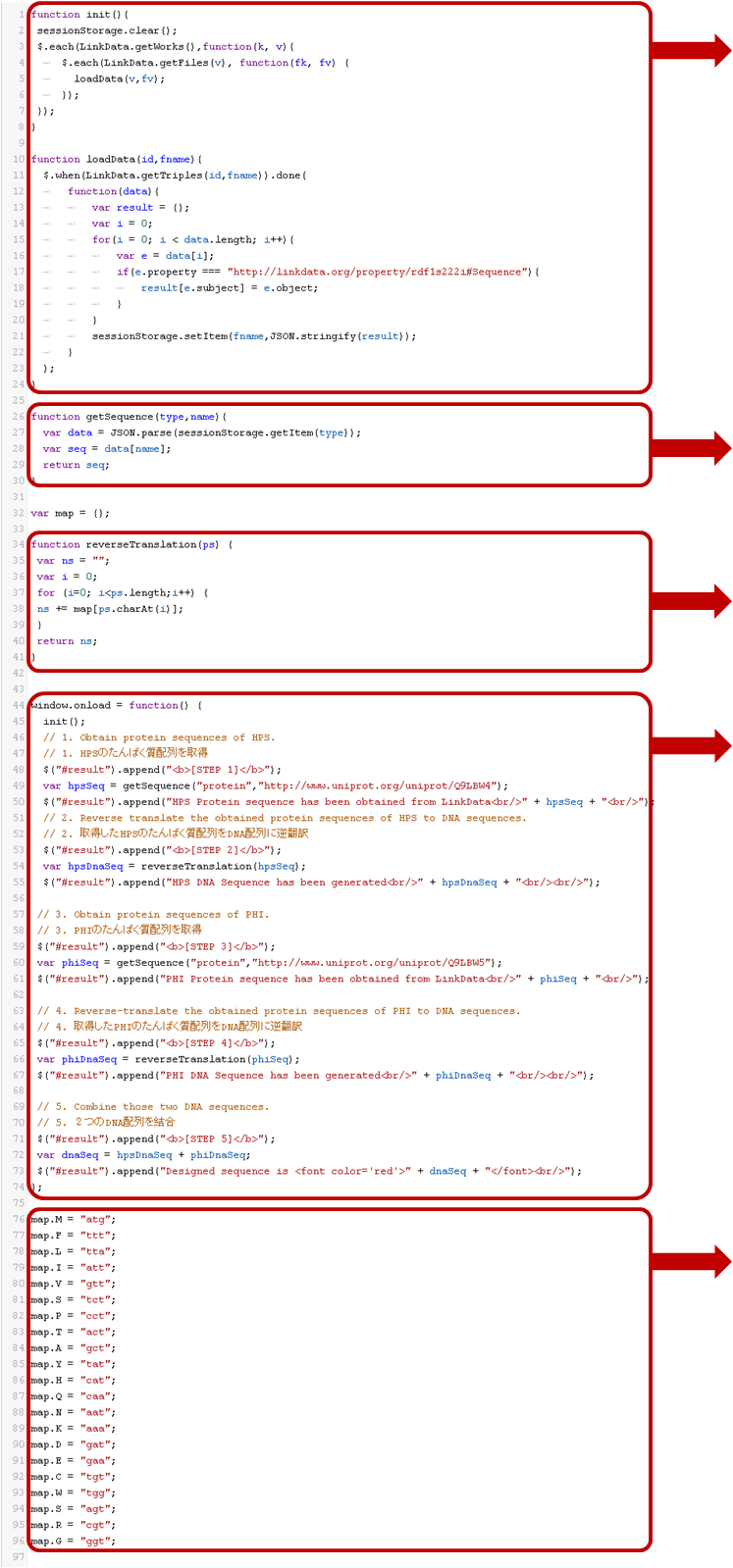
The followings are basic explanation about key parts for Challenge B sample program.
|
Part A. Initialization of the program, data loading
Load the data of 5 tables (dna, promoter, protein, signal_peptide, terminator) included in the sample data (http://linkdata.org/work/rdf1s222i).
Part B. Create a function for getting sequence data
In this part, create a function to get sequences;
We named this function as ‘getSequence’. You can change the function name. Part C. Creation a function for reverse translation In this part, create a function to reverse translate;
We named this function as ‘reverse Translation’. You can change the function name.
Part D. Main Program
Select protein sequences registered in sample data, execute reverse translation by using the function you created in Part C, obtain DNA sequences, outputs of the execution, and display the DNA sequence in the result area.
More detail is explained in the below.
Part E. Define amino acids with the corresponding codons
In this part, amino acids are linked to codons.
*This example is not the best. You may brash up this "map" function considering with codon optimizations to Arabidopsis and/or other idea. *DNA sequences are represented by a letter string such as `ATGC` whereas protein sequences are represented by a letter string such as `MFLIV...`.
|
D-1: Obtain protein sequences
Store protein sequences in a variable named “hpsSeq”.

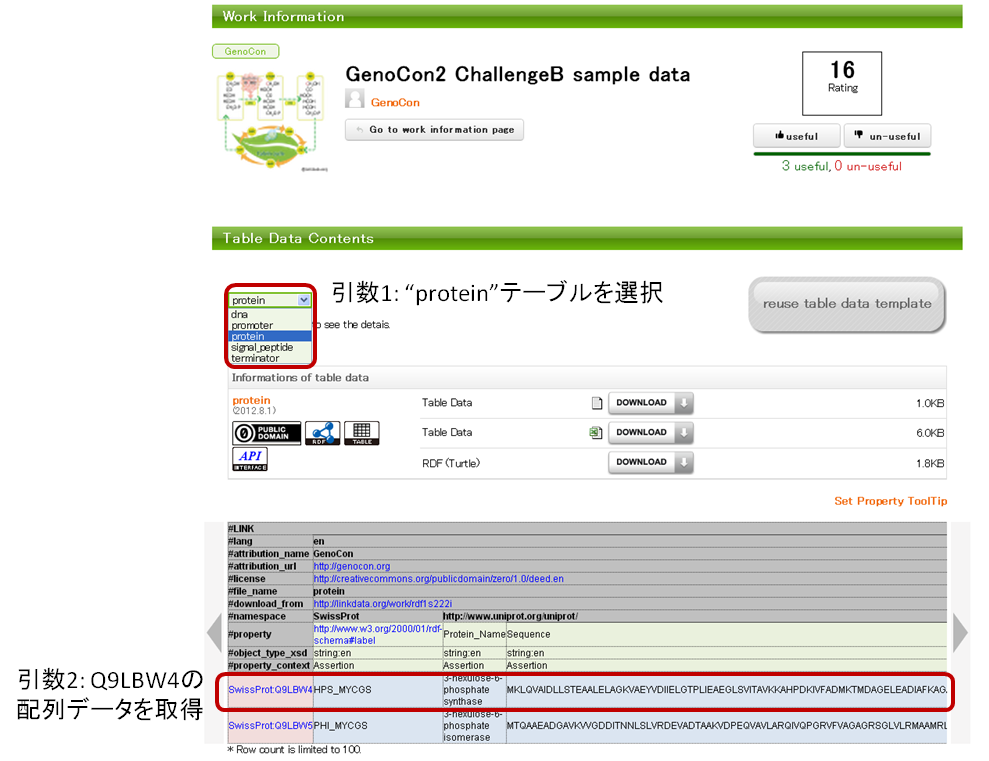
This function selects the "protein" table based on the variable 1, and obtain the sequence data of “Q9LBW4” in the “protein” table based on the variable 2
D-2: Reverse Translation
Translate the protein sequence stored in the variable named "hpsSeq" to a DNA sequence, by using the function created in ③, and store the DNA sequence in variable named "hpsDnaSeq".

D-3: Obtain protein sequences (in a similar way to D-1)

D-4: Reverse translation (in a similar way to D-2)

D-5: Concatenate two DNA sequences
Concatenate two DNA sequences “hpsDnaSeq” (reverse translated in ④-2) and “phiDnaSeq” (reverse translated in ④-4), and store the DNA sequence in variable named “dnaSeq”.

D-6: Display the result
Display the DNA sequence “dnaSeq” in the program executing area.
HTML tags are embedded in the result display area. By appending the strings of DNA sequence into the tag, the design results are displayed.

Expected Results
The plant should get the function to eliminate formaldehyde by inserting two enzymes, HPS (3-hexulose-6-phosphate synthase) and PHI (6-phospho-3-hexuloisomerase) into the Calvin Cycle of the plant.
Importance of This Design
It is known that codon usage is dependent on species. In this design, I reverse transcribed the protein sequences from Mycobacterium gastri to DNA sequences using codons with highest frequency. This is efficient, simple, and easy to understand example of JavaScript coding to construct a DNA sequence created by reverse transcription. There are many possibilities to improve on this design.
References
-
L. M. Chen, H. Yurimoto, K. Z. Li, I. Orita, M. Akita, N. Kato, Y.Sakai, and K. Izui. 2010.
"Assimilation of formaldehyde in transgenic plants due to the introduction of the bacterial ribulose monophosphate pathway genes."
Biosci. Biotechnol. Biochem., 2010. 74:627-635.
-
Mitsui R, Sakai Y, Yasueda H, Kato N. 2000.
"A novel operon encoding formaldehyde fixation: the ribulose monophosphate pathway in the gram-positive facultative methylotrophic bacterium Mycobacterium gastri MB19." J Bacteriol.. 182:944-948.
 Transgenic Arapbidopsis thaliana plant with synthesized DNA
Transgenic Arapbidopsis thaliana plant with synthesized DNA
designed by Finalist participant of the first GenoCon 2010 Challenge
 GenoCon
's
GenoCon Challenge B - The simplest sample
GenoCon
's
GenoCon Challenge B - The simplest sample







